Finite element method getting started examples#
The following page is intended as an example of both the
interaction between
numpyandngsolvein teaching, as well asfor
ngsolvein a Markdown file.
One-dimensional case#
As an introduction to the finite element method, let us consider the scalar boundary value problem
with \(f(x) = 1\).
We can easily obtain the analytical solution in this case. By integrating the right-hand side twice, we obtain a 2nd order polynomial
By inserting the boundary conditions, the analytical solution follows
def uanalytic(x):
return -0.5*x*(x-1)
For the numerical solution, we multiply the differential equation with an arbitrary test function \(v(x)\in C_0^\infty(0,1)\) and integrate over the interval \((0,1)\)
Using partial integration, we obtain the weak equation
The weak problem is given by: Find \(u(x)\) such that \(u(0)=u(1)=0\) and
To fulfill the equation, the solution \(u\) does not necessarily have to be a twice continuously differentiable function. Instead, \(u\) and \(v\) are each differentiated once. It is sufficient to use the functions from the Sobolev space \(H_0^1(0,1)\). The right-hand side also no longer has to be continuous. It is sufficient if the function \(f\) is square integrable, hence \(f\in L_2(0,1)\). From the problem (2) we obtain solutions which do not always necessarily have to be solutions of the strong equation (1).
Let us discretize the interval \((0,1)\) into subintervals \((x_{i-1},x_i)\) for \(i=1,\ldots, n\). On this decomposition we define the basis functions
with which we approximate the Sobolev space \(H_0^1(0,1)\).
Numpy solution#
Show code cell content
import numpy as np
import matplotlib.pyplot as plt
from myst_nb import glue
n=5
a=0
b=1
xi = np.linspace(a,b,n+1)
def phi(i,x,xi=xi):
"""
1d first order FEM basis functions
Parameters
----------
i : int, nummer of the basis function
x : nparray, coordinates to compute
xi : nparray, knots
Return
------
y : nparray, function values for x
"""
y = np.zeros_like(x)
if i > 0:
ind = (xi[i-1]<=x)*(x<=xi[i])
y[ind] = (x[ind]-xi[i-1])/(xi[i]-xi[i-1])
if i < n:
ind = (xi[i]<=x)*(x<xi[i+1])
y[ind] = (xi[i+1]-x[ind])/(xi[i+1]-xi[i])
return y
xp = np.linspace(a,b,400)
fig, ax = plt.subplots(figsize=(6, 2))
for i in range(n+1):
ax.plot(xp,phi(i,xp),label=r'$\varphi_'+str(i)+'$')
ax.legend(loc='upper right', bbox_to_anchor=(1.2, 1))
glue("FEM_1d_p1_fig", fig, display=False)
glue("FEM_1d_p1_n",n,display=False)
In the figure Fig. 2, the interval is divided into 5 subintervals. Accordingly, we have 5 +1 basis functions. Apart from the first and last basis function, the support extends over two subintervals in each case. The basis functions are exactly one in one node \(x_i\), in all others the function value is 0.
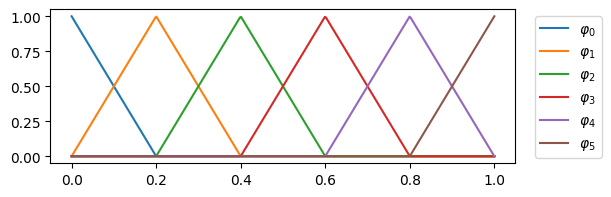
Fig. 2 FEM 1d first order basis functions#
For the weak equation (2) we obtain the finite dimensional problem: Find
such, that \(u(0) = u(1) = 0\) and
Applying \(u_h\) into (3) leads to the system of equation
We define the matrix \(A\) and the vector \(b\)
and obtain the (reduced) linear system of equations for the coefficients \(u_1, \ldots, u_{n-1}\)
or in compact form
with \(\vec{u} = (u_1, \ldots, u_{n-1})^T\) and \(\vec{b} = (b_1, \ldots, b_{n-1})^T\). Due to the Dirichlet boundary conditions we have \(u_0 = u_n = 0\).
Assuming an equidistant decomposition as in the example above Fig. 2, we get
Show code cell content
def dphi(i,x,xi=xi):
"""
Derivative 1d first order FEM basis functionen
Parameters
----------
i : int, nummer of the basis function
x : nparray, coordinates to compute
xi : nparray, knots
Return
------
y : nparray, function values for x
"""
y = np.zeros_like(x)
if i > 0:
h = xi[i]-xi[i-1]
ind = (xi[i-1]<=x)*(x<=xi[i])
y[ind] = np.ones_like(x[ind])/h
if i < n:
h = xi[i+1]-xi[i]
ind = (xi[i]<=x)*(x<xi[i+1])
y[ind] = -np.ones_like(x[ind])/h
return y
fig, ax = plt.subplots(figsize=(6, 2))
for i in range(n+1):
ax.plot(xp,dphi(i,xp),label=r'$\varphi_'+str(i)+'\'(x)$')
ax.legend(loc='upper right', bbox_to_anchor=(1.25, 1.05))
glue("FEM_1d_p1_deriv_fig", fig, display=False)
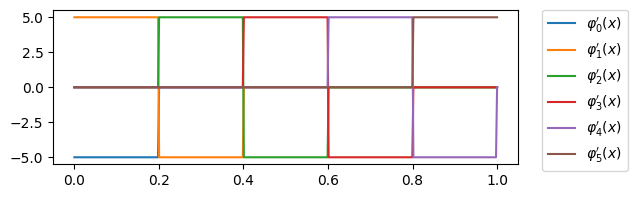
Fig. 3 Derivative 1d first order FEM basis functions#
For the specific example, we obtain the matrix \(A\):
Show code cell source
from scipy.integrate import fixed_quad
A = []
for i in range(0,n+1):
ai = []
for j in range(0,n+1):
# integration over the elements using the Gaussian quadrature
aij = np.sum([fixed_quad(lambda x:dphi(i,np.array(x))*dphi(j,np.array(x)), xi[k], xi[k+1],n=2)[0]
for k in range(n)])
ai.append(aij)
A.append(ai)
A = np.array(A,dtype=float)
A
array([[ 5., -5., 0., 0., 0., 0.],
[-5., 10., -5., 0., 0., 0.],
[ 0., -5., 10., -5., 0., 0.],
[ 0., 0., -5., 10., -5., 0.],
[ 0., 0., 0., -5., 10., -5.],
[ 0., 0., 0., 0., -5., 5.]])
Exercise
Compute the coefficients by hand.
Analog follows for the right-hand side with concrete function \(f(x) = 1\):
Show code cell source
def f(x):
x = np.array(x)
return np.ones_like(x)
b = []
for j in range(0,n+1):
b.append(np.sum([fixed_quad(lambda x:f(np.array(x))*phi(j,np.array(x)),
xi[k], xi[k+1],n=2)[0]
for k in range(n)]))
b = np.array(b,dtype=float)
b
array([0.1, 0.2, 0.2, 0.2, 0.2, 0.1])
The FEM solution \(u(x)\) is therefore given by the solution of the system of equations (6) with the calculated system matrix and right hand side vector. Since the boundary values are given, only the inner degrees of freedom are used. The result is
u = np.zeros_like(xi)
# We will deal with the choice of linear equation solvers later:
u[1:-1] = np.linalg.solve(A[1:-1,1:-1],b[1:-1])
u
array([0. , 0.08, 0.12, 0.12, 0.08, 0. ])
Show code cell content
xp = np.linspace(0,1,400)
fig, ax = plt.subplots(figsize=(6, 2))
ax.plot(xi,u,label='FEM solution')
ax.plot(xp,uanalytic(xp),label='analytical solution')
ax.legend()
glue("FEM_1d_p1_solutionexmp_fig", fig, display=False)
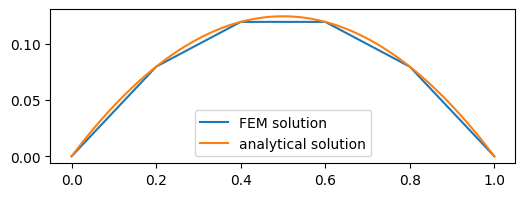
Fig. 4 Solution of the boundary value problem using 1st order FEM#
Exercise
What is the system of equations if the trapezoidal rule is used to integrate the right-hand side?
Calculate the solution using the finite difference method and compare the two systems.
Why does the numerical solution in the nodes match the analytical solution exactly?
NGSolve solution#
Now let’s do the same with NGSolve
First, we create a one-dimensional mesh using
netgen.
from netgen.meshing import Mesh as NGMesh # Caution: Mesh is also available in ngsolve!
from netgen.meshing import MeshPoint, Pnt, Element1D, Element0D
from ngsolve import *
m = NGMesh(dim=1)
# Number of subintervals
N = 5
# Points for the decomposition on the interval [0,1]
pnums = []
for i in range(0, N+1):
pnums.append (m.Add (MeshPoint (Pnt(i/N, 0, 0))))
# Each 1D element (subinterval) can be assigned to a material.
# In our case, there is only one material.
idx = m.AddRegion("material", dim=1)
for i in range(0,N):
m.Add (Element1D ([pnums[i],pnums[i+1]], index=idx))
# Left and right ends are boundary value points (0D elements)
idx_left = m.AddRegion("left", dim=0)
idx_right = m.AddRegion("right", dim=0)
m.Add (Element0D (pnums[0], index=idx_left))
m.Add (Element0D (pnums[N], index=idx_right))
# We have now defined the mesh
mesh = Mesh(m)
Now we create a \(H^1\) function space using this 1D mesh and the Dirichlet boundary points left and right.
V = H1(mesh,order = 1, dirichlet='left|right')
u = V.TrialFunction()
v = V.TestFunction()
# short cut
# u,v = V.TnT()
\(u,v\) are trial and test functions for the definition of the linear and bilinear function
a = BilinearForm(V)
a += grad(u)*grad(v)*dx
f = CoefficientFunction(1)
b = LinearForm(V)
b += f*v*dx
The two operators are now defined, but not yet calculated. The calculation is also called assembling. We will see later what this means in detail.
a.Assemble()
b.Assemble();
The matrix and the vector of the bilinear and linear form contain all degrees of freedom, in particular also the boundary points. However, these are given by the Dirichlet boundary values and do not need to be calculated (as mentioned above). We will take this into account when solving the system.
print(a.mat)
Row 0: 0: 5 1: -5
Row 1: 0: -5 1: 10 2: -5
Row 2: 1: -5 2: 10 3: -5
Row 3: 2: -5 3: 10 4: -5
Row 4: 3: -5 4: 10 5: -5
Row 5: 4: -5 5: 5
print(b.vec)
0.1
0.2
0.2
0.2
0.2
0.1
The GridFunction class is available in NGSolve for the solution itself. The trial and test functions are only used to define the problem and therefore have no memory for the solution vector, for example. The linear combination of the basis functions is automatically calculated in the evaluation of this. We need a GridFunction for the solution:
gfu = GridFunction(V)
Calculation of the FEM solution with NGSolve taking into account the free degrees of freedom:
gfu.vec.data = a.mat.Inverse(freedofs=V.FreeDofs())*b.vec
When the command is executed, the matrix is not inverted and multiplied to the vector from the left. That would be far too time-consuming numerically. Even if the notation claims otherwise, only the system of equations is solved.
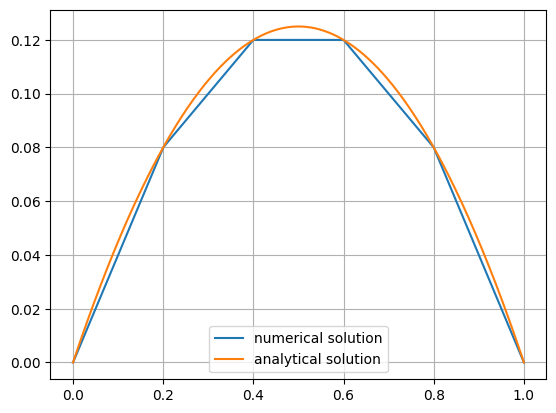
Again, we get the same result as above:
Show code cell source
xp = np.linspace(0,1,400)
plt.plot(xp,[gfu(mesh(xi,0)) for xi in xp],label='numerical solution')
plt.plot(xp, uanalytic(xp),label='analytical solution')
plt.legend()
plt.grid()
plt.show()
Extension into two-dimensional space#
Now let us look at the two-dimensional case. We will see that apart from the mesh, hence the region, the implementation in ngsolve remains identical. We consider the analog boundary value problem in two dimensions
on the unit square in \(\mathbb{R}^2\). For the weak equation follows
where \(V\) is a finite element space with \(\dim V < \infty\).
from ngsolve import *
from ngsolve.webgui import Draw
From netgen we import the unit square. With the help of the netgen module we can define and mesh 2d and 3d geometries.
from netgen.geom2d import unit_square
We now generate an unstructured mesh with a maximum edge length of 0.25:
mesh = Mesh(unit_square.GenerateMesh(maxh=0.25))
The size of the mesh can be controlled via the maxh parameter.
Draw(mesh);
What do the basis functions look like in two dimensions? To do this, we create an FEM function space and visualize the basis functions. The edge of the unit square consists of 4 lines, which have different labels:
mesh.GetBoundaries()
('bottom', 'right', 'top', 'left')
We define the H1 FEM function space with the Dirichlet boundary condition and initialize a GridFunction, an FEM function from the function space:
V = H1(mesh,order=1,dirichlet='bottom|right|top|left')
gfu = GridFunction(V)
Using the grid function, we can visualize the global basic functions. To do this, we set one coefficient to 1 and the others to 0. In the following example we visualize the 20th global basis function:
gfu.vec.FV()[:] = 0 # Initialize all entries with 0
gfu.vec.FV()[20] = 1
Draw(gfu,mesh,'u');
Using \(u(x) = \sum_i u_i\,\varphi_i(x) \in V\), as in the 1d example (4), the same system of equations follows
with the bilinear form \(A: V \times V \to \mathbb{R}\)
and linear form \(f: V \to \mathbb{R}\)
For the definition in ngsolve we use again the proxy functions
u = V.TrialFunction()
v = V.TestFunction()
In two dimensions, we use dx to denote a surface integral and ds for a path integral (boundary integral). In our case we have for the bilinear form
A = BilinearForm(V)
A += grad(u) * grad(v)*dx
and analogously for the linear form
f = CoefficientFunction(1)
b = LinearForm(V)
b += f*v*dx
The right-hand side function onece again defined using a CoefficientFunction.
We have thus defined the bilinear form, which can be described in finite dimensions using a matrix, and the linear form, which can be described using a vector, but have not yet calculated them. The calculation of these is also called assembling.
A.Assemble()
b.Assemble();
The matrix \(A\) is in a sparse format stored. This means that only the matrix entries for which we potentially receive an entry are saved. The memory is not allocated at all for the rest of the matrix.
print(A.mat)
Row 0: 0: 1 4: -0.5 15: -0.5
Row 1: 1: 0.895075 6: -0.377531 7: -0.378808 18: -0.138736
Row 2: 2: 1 9: -0.5 10: -0.5
Row 3: 3: 1 12: -0.5 13: -0.5
Row 4: 0: -0.5 4: 1.92156 5: -0.31533 15: -0.15374 16: -0.95249
Row 5: 4: -0.31533 5: 1.8612 6: -0.198542 16: -0.307165 17: -1.04016
Row 6: 1: -0.377531 5: -0.198542 6: 1.79993 17: -0.395887 18: -0.827968
Row 7: 1: -0.378808 7: 1.84286 8: -0.324152 18: -0.918526 19: -0.22137
Row 8: 7: -0.324152 8: 1.79252 9: -0.271716 19: -0.860521 20: -0.336129
Row 9: 2: -0.5 8: -0.271716 9: 2.02128 10: -0.0759864 20: -1.17357
Row 10: 2: -0.5 9: -0.0759864 10: 1.81976 11: -0.220817 20: -0.288822 21: -0.734131
Row 11: 10: -0.220817 11: 1.84084 12: -0.448101 21: -0.875576 22: -0.296345
Row 12: 3: -0.5 11: -0.448101 12: 1.87 13: -0.26789 22: -0.654007
Row 13: 3: -0.5 12: -0.26789 13: 1.84321 14: -0.506823 22: -0.367252 23: -0.201248
Row 14: 13: -0.506823 14: 2.01367 15: -0.420679 23: -1.08617
Row 15: 0: -0.5 4: -0.15374 14: -0.420679 15: 1.79625 16: -0.418051 23: -0.303778
Row 16: 4: -0.95249 5: -0.307165 15: -0.418051 16: 3.61084 17: -0.609253 23: -0.717205 24: -0.606678
Row 17: 5: -1.04016 6: -0.395887 16: -0.609253 17: 3.78711 18: -0.965502 24: -0.77631
Row 18: 1: -0.138736 6: -0.827968 7: -0.918526 17: -0.965502 18: 3.86126 19: -0.936881 24: -0.073651
Row 19: 7: -0.22137 8: -0.860521 18: -0.936881 19: 3.63019 20: -0.534253 24: -0.503238 25: -0.573927
Row 20: 8: -0.336129 9: -1.17357 10: -0.288822 19: -0.534253 20: 3.74798 21: -0.678509 25: -0.73669
Row 21: 10: -0.734131 11: -0.875576 20: -0.678509 21: 3.72565 22: -0.616041 25: -0.821391
Row 22: 11: -0.296345 12: -0.654007 13: -0.367252 21: -0.616041 22: 3.58077 23: -0.913832 24: -0.139859 25: -0.593436
Row 23: 13: -0.201248 14: -1.08617 15: -0.303778 16: -0.717205 22: -0.913832 23: 3.76762 24: -0.545392
Row 24: 16: -0.606678 17: -0.77631 18: -0.073651 19: -0.503238 22: -0.139859 23: -0.545392 24: 3.67338 25: -1.02825
Row 25: 19: -0.573927 20: -0.73669 21: -0.821391 22: -0.593436 24: -1.02825 25: 3.7537
We can also regard this matrix (at least as long as it is small!) as a dense matrix:
rows,cols,vals = A.mat.COO()
denseA = np.zeros((np.max(rows)+1,np.max(rows)+1))
k=0
for i,j in zip(rows,cols):
denseA[i,j] = vals[k]
k+=1
plt.spy(denseA)
plt.show()
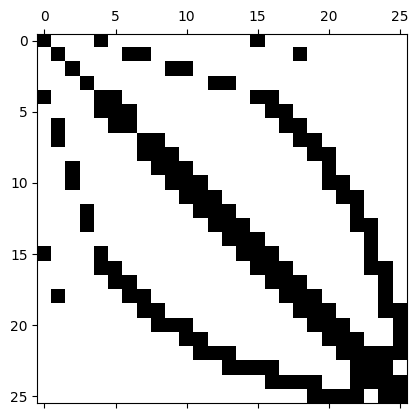
We can also use scipy.sparse for further computations.
import scipy.sparse as sp
Asparse = sp.coo_matrix((vals,(rows,cols)))
plt.spy(Asparse)
<matplotlib.lines.Line2D at 0x10ee2f250>
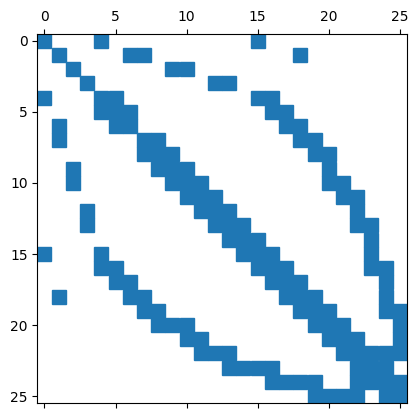
The pattern of the matrix entries depends on the numbering of the nodes (from the meshing) and the FEM basic functions. It is also possible to draw the mesh directly with matplotlib; the mesh object contains all the information required for this.
for e in mesh.edges:
line = np.array([mesh.vertices[v.nr].point for v in e.vertices])
plt.plot(line[:,0],line[:,1],c='gray',alpha=0.75)
for v in mesh.vertices:
plt.text(*v.point,v,color='red')
plt.gca().set_axis_off()
plt.gca().set_aspect(1)
plt.show()
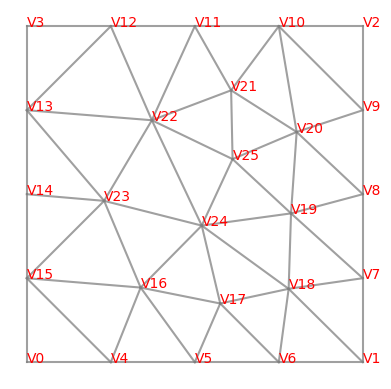
We now consider the free degrees of freedom, for which we must keep the homogeneous Dirichlet boundary condition in mind. The number of free dof’s is given by
freedofs = V.FreeDofs()
print(freedofs)
freedofsnp = np.array([i for i in freedofs])
0: 00000000000000001111111111
As we can see, the order of numbering the nodes is via the corners, edges into the interior. Thus, the degrees of freedom available to us are at the end. The matrix pattern for solving the problem is given by
ind = np.arange(freedofsnp.shape[0])[freedofsnp]
plt.spy(denseA[np.ix_(ind,ind)])
plt.show()
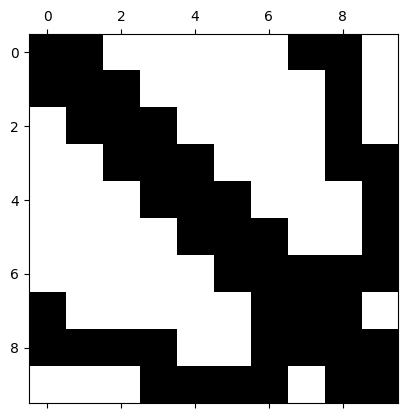
Let’s solve the system for the inner degrees of freedom:
gfu.vec.data = A.mat.Inverse(freedofs=V.FreeDofs())*b.vec
Draw(gfu,mesh,'u');
